Blast was developed to work on Mac OS X 10.6.6 or later. The program's installation file is generally known as blast.zip. This Mac application is a product of baKno. OAKLAND - The suspected leader of a notorious West Oakland street gang was found guilty Thursday of seven felonies, including attempted murder, and now faces at least 40 years in state prison.
but keeps all your important events and tasks always visible on your desktop.
Events and Tasks
Rainlendar supports events and tasks which both are kept in separate lists. This helps you to keep your life better organized and makes it easier to see what are the upcoming things you need to do.
Alarms
Get notified in advanced before the event is due so that you don't forget your important events. It is also possible to snooze the alarm if you want to get reminded about it later.
Standards Based
All the data is stored in the standard iCalendar format (RFC2445) which is supported by most calendar applications. This makes it easy to transfer the events between applications.
Highly Customizable
The look and feel of the calendar can be changed with skins. You can also mix the skins together and have as many windows visible as you want.
Localized
Rainlendar has been translated to over 50 different languages so you can use it in your native language. It's also possible to use any language when entering the events.
Platform Independent
The application works in all major operating systems: Windows, Mac OS X and Linux. All the data, skins, languages and scripts are the same in all platforms so you can easily migrate between them.
Shared & 3rd Party Calendars
With Rainlendar Pro you can subscribe to online calendars and share your events with other computers. You can also see and edit your Outlook appointments and tasks in Rainlendar (Windows only).
Step 1: Introduction
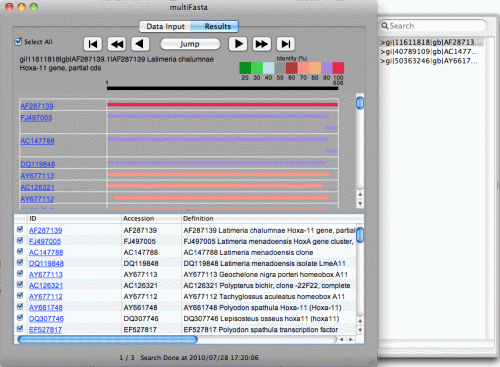
This tutorial will take you through the steps involved in looking for an alignment between two DNA sequences. It uses the Find Overlaps tool, which is designed to look for dove-tail (end-to-end) alignments.
This tutorial assumes the user has already reviewed at least Basic Operation tutorial and has a basic knowledge of the program.
We will use Genome Workbench to review alignments and how to visually inspect annotations on two sequences in an alignment.
Step 2: Selecting Sequences to Align
For this exercise we selected two clone sequences (AC040978.8, AC115836.5) that are parts of the tiling path track for Human chromosome 8 (NC_000008) and are known to have overlap. First let us add both sequences into a new Genome Workbench project. Click File => Open, in dialog select Data from GenBank and paste AC040978.8, AC115836.5 into the Accession to Load box, click Next and Finish. Note: the other way to add data to the project is to use Search View (see Basic Operation tutorial, Step 3).
Now there is a 'New Project' in the Project Tree View. Let us create a folder called Sequences in our project and add both sequences to this folder.
Right click (or control click on the Mac OS) on the 'Data' label and choose 'New Folder' from the contextual menu. In the New Project Folder dialog that appears, names the folder Sequences.
Now let us put our sequences in the Sequences folder. This can be done by selecting the sequences, right clicking and choosing Cut, then selecting the Sequences folder, right clicking, and choosing Paste. The other way is just to select the sequences and move them using drag and drop functionality.
You can rename projects by selecting the project (green folder named 'New Project' in the project tree), right-clicking, and selecting Properties. Rename this project to: AC040978.8_AC115836.5_alignment.
Step 3: Generating an Alignment
We will generate a specialized alignment for these two sequences - an overlap alignment. This alignment expresses the relationship seen between two clones assembled sequentially in a tiled BAC assembly.
Select both sequences within the project tree. Then select Tools=> Run Tool from the main menu. In the Run Tool dialog, choose Find Overlap between DNA Sequences and click Next.
In the dialog that opens, select AC040978.8 as Sequence 1 and AC115836.5 as Sequence 2. Use the default alignment parameters for this alignment and select Next.
Then click Finish. When the alignment is complete, the result will appear in the AC040978.8_AC115836.5_alignment project in the Tool Result folder.
Note: The Find Overlaps tool first looks for a BLAST alignment between the component sequences, and if none is found, goes on to perform a banded global alignment. As the alignment is performed locally, repeat filtering is only available to external users if the repeats for the components have also been loaded locally.
Step 4: Viewing the Alignment
To view this alignment right click on it in the Project View and choose Open New View in the context menu. From the Open View dialog choose Multi-pane Cross Alignment View. You will see a view like the one shown below. This viewer packs three views together - a Dot Matrix view of the alignment, and two Graphical Views, one on each sequence. Each view can be resized by clicking and dragging on its edges. Resize the panels to match the image shown. In this composite view, you should see a dot matrix view on left and two graphical views stacked vertically on the right.
The dot matrix view shows a single diagonal line that represents the alignment. Sequence 1 (AC040978.8) is on the Y-axis and Sequence 2 (AC115836.5) is on the X- axis. The negative slope of the line indicates the two sequences align in opposing orientations.
The top graphical window shows Sequence 1 as the master (anchor) sequence with Sequence 2 aligned beneath. In contrast, the bottom graphical window shows Sequence 2 as the master (anchor) sequence with Sequence 1 aligned beneath.
Depending on your personal track configuration, you might see a bit different set of tracks in the graphical views on the right. In our example we show three tracks: master (anchor) sequence, alignment track and SNP (variation) track. If there are other features annotated on the master (anchor) sequences, they are available to see in the graphical views as separated tracks. To reveal/hide available tracks, use the Context icon at the bottom or the Gear icon on the upper panel in both graphical windows (see Basic Operation tutorial for more information).
Blast Lander Mac Os Catalina
A tooltip (pop-up window) containing additional information will open when the mouse is held over any alignment or feature. In the image below tooltip shows alignment information including percent of coverage and identity, mismatches, gaps and unaligned regions.
Step 5: Taking a closer look

Blast Lander Mac Os Catalina
A tooltip (pop-up window) containing additional information will open when the mouse is held over any alignment or feature. In the image below tooltip shows alignment information including percent of coverage and identity, mismatches, gaps and unaligned regions.
Step 5: Taking a closer look
For a closer view of the alignment, double click on the gray alignment bar in either graphical views of Multi-pane view. This action will select alignment as an object and zoom to the level of the alignment in both graphical views and highlight selected alignment in the dot matrix view.
If you select region in the one of the graphical views it is also highlighted in the dot matrix view and vise versa.
Marks within the alignment bars in the graphical views indicate mismatches, insertions and gaps. You can see these alignment details by increasing your zoom. In this example, zooming in on the lower graphical window reveals two base pair indel in the alignment at Sequence 2 (AC115836.5) position 177,420. A known variation associated with it is also shown.
Step 6: Additional Views: Alignment Span View
The Alignment Span Table View provides information about each segment of an alignment. In the case of a discontinuous alignment (such as the one in this tutorial), information about each segment of the alignment is represented in its own row. To see this view, select the alignment icon in the Project Tree and choose View=>Open View and choose Alignment Span View from the dialog. Or right click on the alignment and choose Open View and then choose Alignment Span View from the dialog. (Note: you can manage merging window size using right-click in the table and select Settings and set the threshold for merging window to a smaller size).
Broadcasting option can be used between span and multi-pan views. Select row in the Span Alignment Table View and jump to the Multi-pane Cross Alignment View, observe that regions selected in the span view are selected in all windows in the multi-pan view.
Step 7: Exporting Alignments
Alignments can be exported from Genome Workbench in a couple of formats. For the purposes of submission to NCBI, alignments should be exported in ASN.1 format.
In the graphical view, select an alignment and choose File=> Export from the main menu. A dialog box will open. From the list menu on the left side of this box, select ASN File. Choose one or more of the alignments (control (or shift) click for multiple selection). Choose a file name by clicking the small box labeled ... to select the file destination. Select Text for the ASN type and click Finish.
Step 8: Finished
Blast Lander Mac Os 11
Congratulations! You now know how to perform a basic alignment between two DNA sequences in order to find a dove-tail overlap. You have also learned several ways to view alignments, and how to export an alignment from Genome Workbench.
Current Version is 3.6.0 (released March 04, 2021)

